Breast Cancer Malignancy Diagnostics using Neural Networks
This project applies statistical learning techniques and neural networks to classify breast cancer malignancy based on cellular nuclei measurements. By comparing traditional classification methods with deep learning approaches, the goal was to improve diagnosis accuracy and reduce human error in cytological inspection.
Project Goals
- Build and compare multiple models (including classical classifiers and neural networks) for breast cancer diagnosis.
- Focus on feature transformation (squaring, logarithmic) to improve classification accuracy.
- Find the optimal deep learning architecture for the task.
Challenges and Solutions
Challenge: Feature selection and model architecture optimization.
Solution:
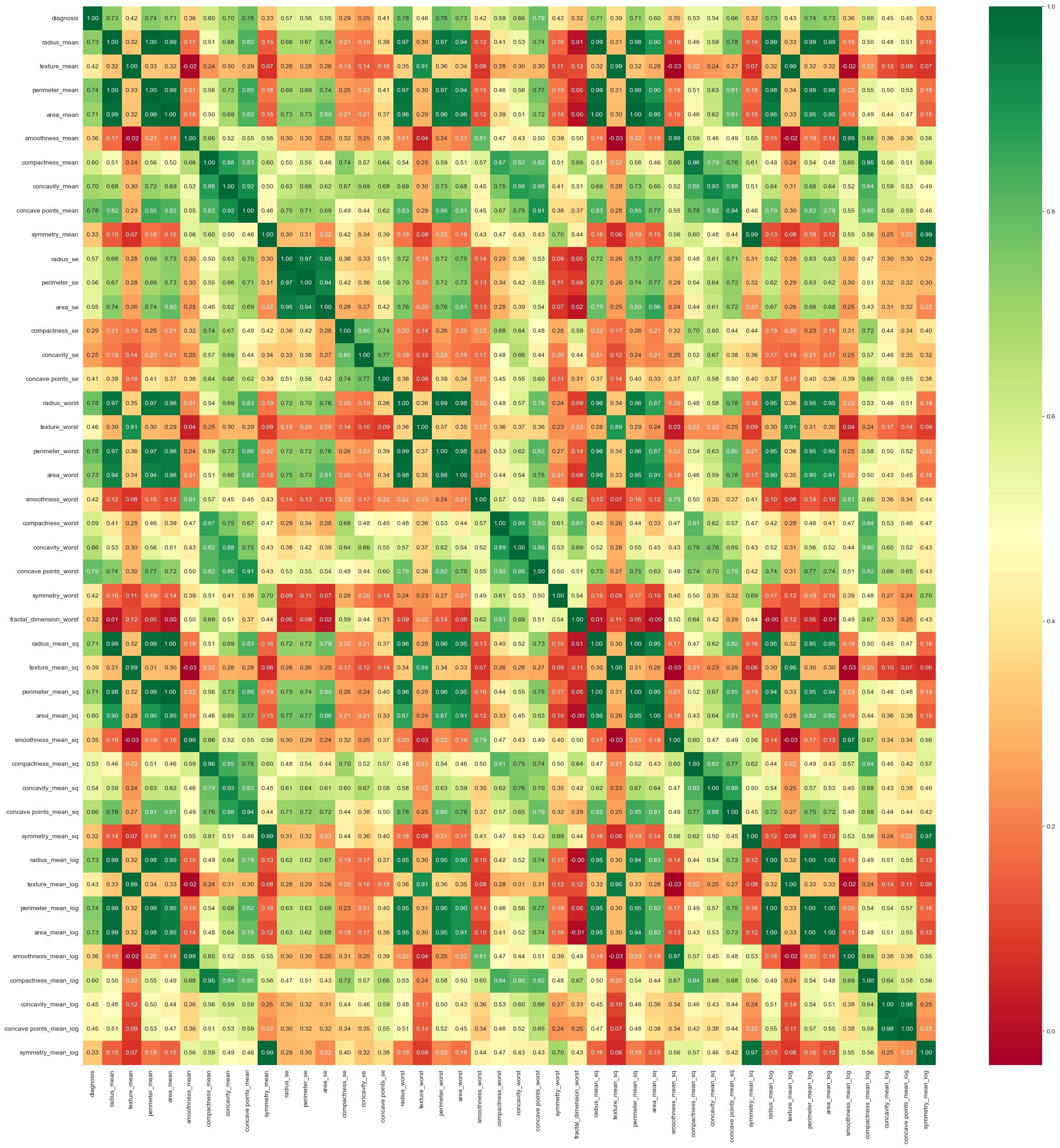
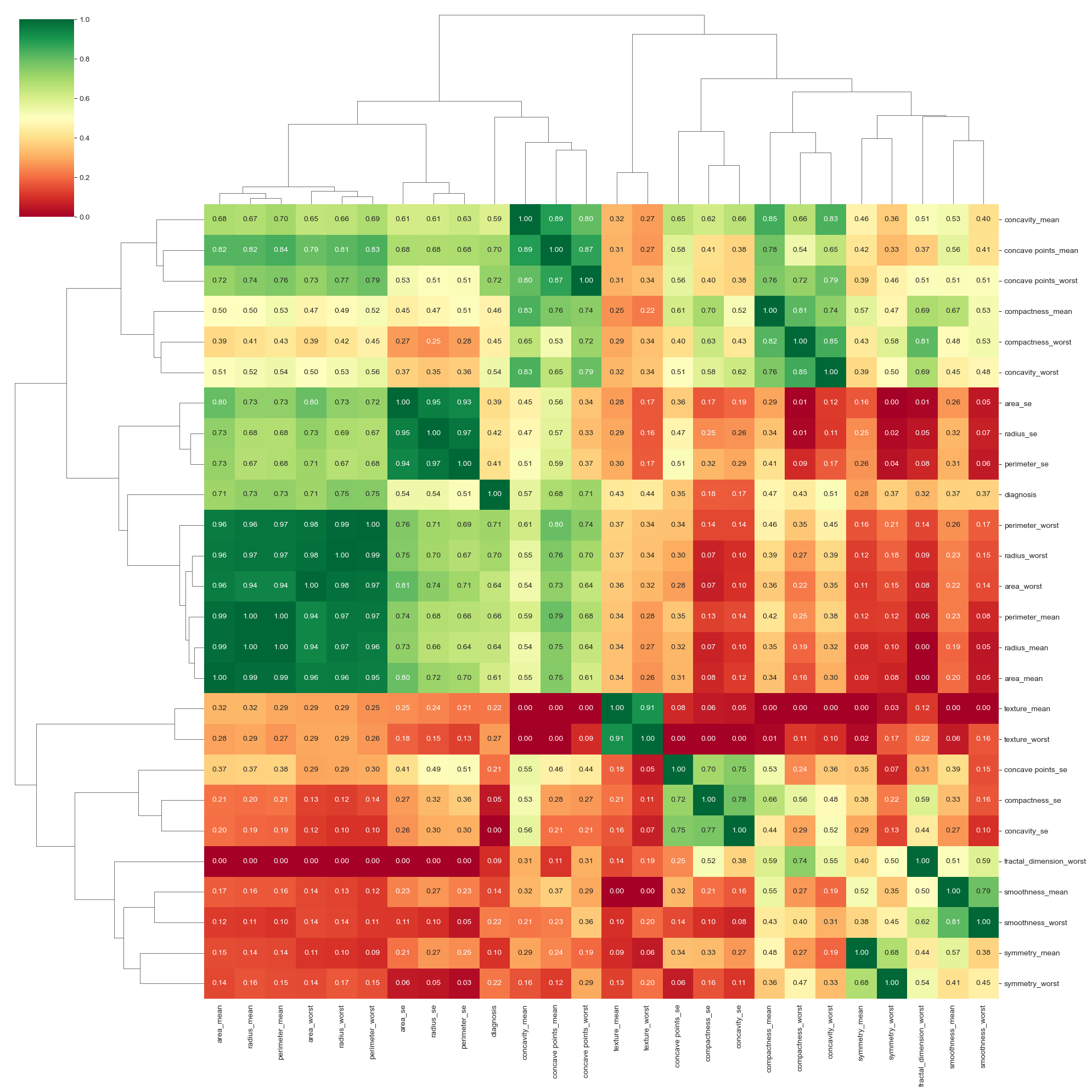
- Eliminated features with correlations below 40% to diagnosis.
- Applied transformations (feature squaring, logarithmic) to enhance model inputs.
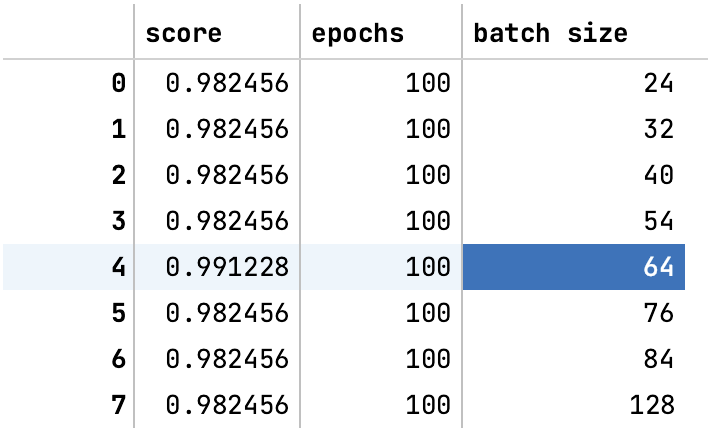
- Used grid search and parameter tuning to optimize neural network performance.
Technologies and Tools Used
Languages/Technologies: Python, TensorFlow, Scikit-Learn
Libraries: Pandas, Seaborn, Matplotlib
Modeling: Neural Networks, Random Forest, KNN, SVM
Other Tools: Grid Search, Feature Engineering
Data Description
Dataset: Breast Cancer Wisconsin Diagnostic dataset from Kaggle (569 samples).
Features: Ten cellular nuclei measurements for each sample (radius, texture, perimeter, area, etc.), along with standard error and "worst" values for certain features.
Feature Removal: Five features with correlations below 40% were removed, resulting in 33 features for the final model.
Methodology
Data Preprocessing: Removed low-correlation features, applied MinMax scaling, created squared and log-transformed versions of the data.
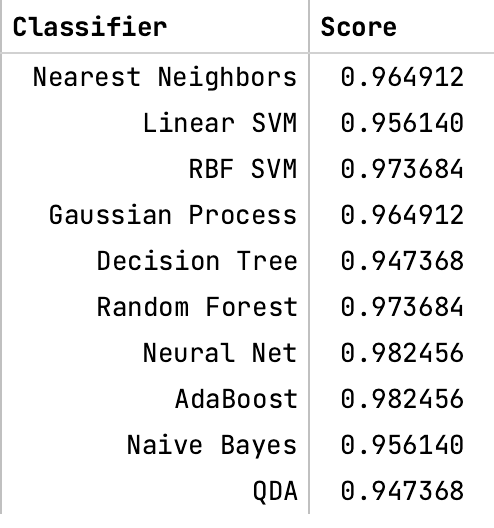
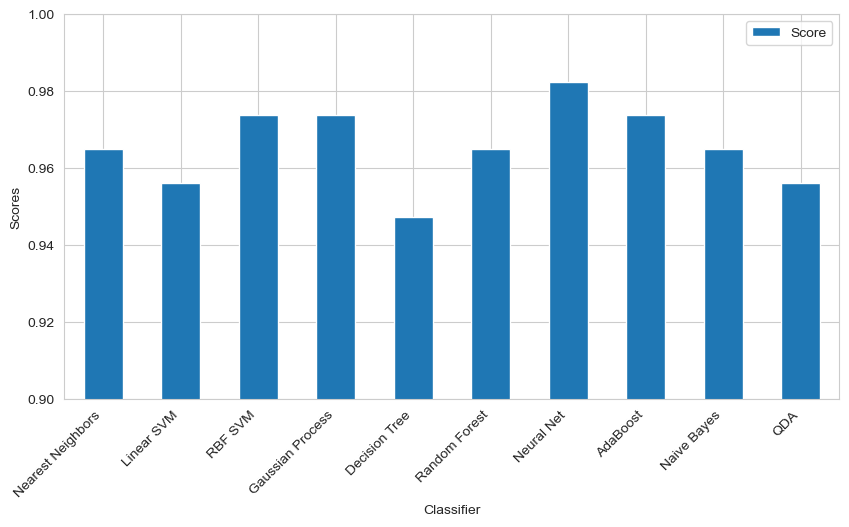
Models Compared: Classifiers included KNN, SVM (Linear/RBF), Random Forest, Decision Trees, Neural Networks, etc.
Neural Network Architecture: 4 hidden layers, 33-24-36 neurons, ReLU activations, final layer with sigmoid activation.
Key Results
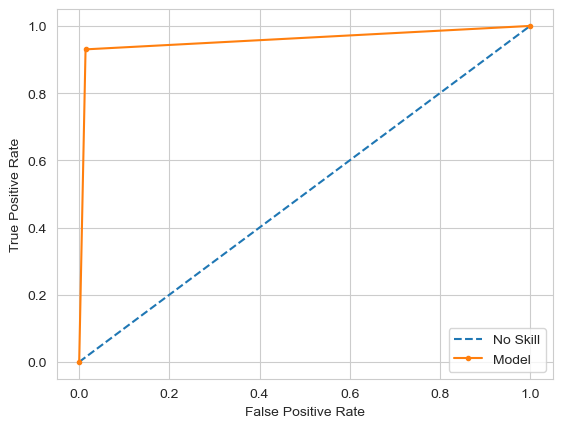
- The deep neural network achieved the highest accuracy of 98.25%, outperforming classical methods.
- Key Features for Diagnosis: Concave Points Worst (most significant factor in differentiating between malignant and benign), Mean Area, and Texture.
Data Visualization
Analysis & Interpretation
The deep neural network model achieved high accuracy, but classical methods like Random Forests and KNN performed comparably. Concave points and area-related features were the most significant factors in diagnosing malignancy. Further hyperparameter tuning could offer marginal improvements, but the current model architecture is near optimal.
Conclusion
The deep neural network outperforms traditional models, offering a balanced trade-off between complexity and diagnostic accuracy. Future work could involve exploring more complex feature engineering, outlier detection, and advanced hyperparameter tuning.
Screenshots